How to cut costs in my lab
I've been on a role the last couple of days. However, I must admit, I got a bit carried away with a couple of long screeds. So last night I took a break. Today I'll post something that I had originally written last week to be posted on Friday. The reason it was never posted is because it was pre-empted by my post about David Irving and my taking more time than I should have wasted slapping down Dean Esmay's ridiculous HIV denialism on Monday. On to lighter fare...
Last week, via Science and Politics, I think I might have found a desperately needed way to cut costs in my lab: The MacGyver Project: Genomic DNA Extraction and Gel Electrophoresis Experiments Using Everyday Materials. Such cost cutting measures are critical, because, despite getting my first NIH grant earlier this year, finances are still tight. Oh, they were fine this year, and you'd think I'd be be riding high in hog heaven (scientifically and budgetarily speaking), after finally having attained the Holy Grail of biomedical research funding: the NIH R01. Unfortunately, the lab isn't. First off, the NIH took 22% off the top of all new grants with the rather scary possibility of additional cuts to existing grants if the NIH budget takes a cut in the 2006 budget, as is a distinct possibility. Second, my other grant is due to expire in April, after which money will get much tighter. Third, the university mandates that salaries go up (regardless of employee merit) 4% a year, and the NIH has been consistently raising the mandated rates for postdocs and graduates students every year. In addition, the university has been steadily increasing the amount they charge for fringe benefits and taxes. When I first arrived 6 years ago, it was 26% of salary. Last year, it was 33%, and they plan on increasing it to 34% for 2006. Salaries and fringes keep going up, but the grant provides the same sum every year.
There are only two possible solutions (which may be used in combination, of course): Raise revenues (i. e., get another grant, something I'm working on); or cut expenses. Salaries are pretty much untouchable; so that just leaves supplies.
And that's where the MacGyver Project comes in.
For those of you not familiar with MacGyver, it was a TV show from the late 1980's in which the hero, MacGyver was able to solve problems and defeat his enemies through his cleverness and ability to improvise. His clever solution to problems using the materials at hand, such as using Rosary beads to focus sunlight and trigger a slingshot or using acetic acid and ammonia to produce a smoke screen, became known as MacGyverisms.
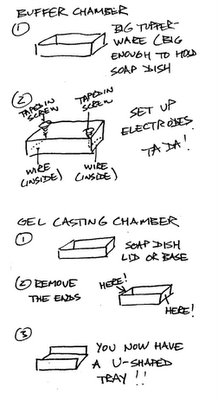
So, in the spirit of MacGyver, why waste all that money buying gel electrophoresis boxes at $250 a pop for what is basically a Plexiglass box with a couple of wires and plugs and a tray to run the gel in? What a rip-off! And why waste hundreds of dollars a month on various DNA purification kits, when much cheaper methods will do? I could totally see using Agar Agar obtained from an Asian grocery store (and, conveniently, there is a large such store a mere four miles or so away). Legos with Glad Press 'N' Seal to construct the gel tray? Genius! See the diagram:
Unfortunately, we don't do genomic DNA extractions very often; so this probably wouldn't be much use. What I really need is a good MacGyver method to extract RNA for quantitative reverse transcriptase real time PCR. We do a lot of quantitative RT-PCR in our lab. A lot. One problem with the MacGyver method is that it would be hard to keep RNases from degrading the RNA. DNA is a lot hardier than RNA, which is easily degraded. It would also be nice to have a MacGyver protocol for the extraction of protein for Western blots and nuclear extracts for gel shift assays.
I suppose it would be too much to hope for a MacGyver protocol for cloning, plasmid isolation, and retroviral vector construction--or for the PCR reaction itself. For one thing, you can't do PCR with Taq polymerase or other thermostable DNA polymerases, and the only sort of MacGyver protocol I can think of would involve harvesting the thermophilic bacteria and somehow isolating the enzyme.
Still running a lab on the MacGyver concept sounds as though it would be rather fun. Unfortunately, it would not be practical.
Last week, via Science and Politics, I think I might have found a desperately needed way to cut costs in my lab: The MacGyver Project: Genomic DNA Extraction and Gel Electrophoresis Experiments Using Everyday Materials. Such cost cutting measures are critical, because, despite getting my first NIH grant earlier this year, finances are still tight. Oh, they were fine this year, and you'd think I'd be be riding high in hog heaven (scientifically and budgetarily speaking), after finally having attained the Holy Grail of biomedical research funding: the NIH R01. Unfortunately, the lab isn't. First off, the NIH took 22% off the top of all new grants with the rather scary possibility of additional cuts to existing grants if the NIH budget takes a cut in the 2006 budget, as is a distinct possibility. Second, my other grant is due to expire in April, after which money will get much tighter. Third, the university mandates that salaries go up (regardless of employee merit) 4% a year, and the NIH has been consistently raising the mandated rates for postdocs and graduates students every year. In addition, the university has been steadily increasing the amount they charge for fringe benefits and taxes. When I first arrived 6 years ago, it was 26% of salary. Last year, it was 33%, and they plan on increasing it to 34% for 2006. Salaries and fringes keep going up, but the grant provides the same sum every year.
There are only two possible solutions (which may be used in combination, of course): Raise revenues (i. e., get another grant, something I'm working on); or cut expenses. Salaries are pretty much untouchable; so that just leaves supplies.
And that's where the MacGyver Project comes in.
For those of you not familiar with MacGyver, it was a TV show from the late 1980's in which the hero, MacGyver was able to solve problems and defeat his enemies through his cleverness and ability to improvise. His clever solution to problems using the materials at hand, such as using Rosary beads to focus sunlight and trigger a slingshot or using acetic acid and ammonia to produce a smoke screen, became known as MacGyverisms.
So, in the spirit of MacGyver, why waste all that money buying gel electrophoresis boxes at $250 a pop for what is basically a Plexiglass box with a couple of wires and plugs and a tray to run the gel in? What a rip-off! And why waste hundreds of dollars a month on various DNA purification kits, when much cheaper methods will do? I could totally see using Agar Agar obtained from an Asian grocery store (and, conveniently, there is a large such store a mere four miles or so away). Legos with Glad Press 'N' Seal to construct the gel tray? Genius! See the diagram:
Unfortunately, we don't do genomic DNA extractions very often; so this probably wouldn't be much use. What I really need is a good MacGyver method to extract RNA for quantitative reverse transcriptase real time PCR. We do a lot of quantitative RT-PCR in our lab. A lot. One problem with the MacGyver method is that it would be hard to keep RNases from degrading the RNA. DNA is a lot hardier than RNA, which is easily degraded. It would also be nice to have a MacGyver protocol for the extraction of protein for Western blots and nuclear extracts for gel shift assays.
I suppose it would be too much to hope for a MacGyver protocol for cloning, plasmid isolation, and retroviral vector construction--or for the PCR reaction itself. For one thing, you can't do PCR with Taq polymerase or other thermostable DNA polymerases, and the only sort of MacGyver protocol I can think of would involve harvesting the thermophilic bacteria and somehow isolating the enzyme.
Still running a lab on the MacGyver concept sounds as though it would be rather fun. Unfortunately, it would not be practical.

There was a time when *most* scientists built their own equipment. Unfortunately, that costs you in repeatability and probably reliability. And much modern equipment really does depend on a *serious* high-tech/industrial base!
ReplyDeleteOn the other hand, if all the labs in your university pooled their resources, they possibly could save a fair bit by self-manufacturing some of the simpler and more common stuff.
I suppose it would be too much to hope for a MacGyver protocol for cloning, plasmid isolation, and retroviral vector construction--or for the PCR reaction itself. For one thing, you can't do PCR with Taq polymerase or other thermostable DNA polymerases, and the only sort of MacGyver protocol I can think of would involve harvesting the thermophilic bacteria and somehow isolating the enzyme.
ReplyDeleteI've heard of labs growing their own Taq bacteria, and isolating the DNA polymerase. The problem is that you really can't get much polymerase without industrial size populations and industrial techniques. I don't think you can break even unless you're doing tens of thousands of reactions a day. You could probably also synthesize your own nucleotides for PCR, but it wouldn't be worth it.
I've heard rumours in the lab about "Kitchen Taq". It's not so much a MacGyverism as it is patent violation.
ReplyDelete1. Buy a tiny quantity of Taq polymerase from your usual supplier
2. Buy lots of Taq polymerase illegally from your colleague down the hall who neglected to clean out his water bath for 10 years
3. Dilute the legal Taq to homeopathic concentrations
4. Do PCR normally, using the illegal Taq with a microlitre or less of super-dilute legal Taq added
5. Write up your results, and claim with a straight face that you used real Taq from the respected, legal supplier
6. Funnel the money you saved into annonymous offshore accounts
Actually cheap (basically free, but slightly illegal) Taq is possible. There exists a "pTaq" protocol (you can google this I believe, and it was once published in an issue of Biotechniques), which essentially uses the native sequence for the heat stable polymerase (the main reason why I'm vague on the legality since most commercially available Taq has been modified to some extent) in a bacterial protein expression vector. What's great about this, is that it's possible to get your Taq, by simply growing up the e.coli. Purification? Resuspend bug pellet in a buffer, boil and spin down debris. It basically takes advantage of the fact that the only thing surviving this process is the Taq itself.
ReplyDeleteOf the record (if there is such a thing in this manner of communication), when we tried it, it worked great. The catch, of course, is that whilst this works well in a bind (i.e. in our case, we're exploring this as an option in developing countries - the thermal cycler is very easy to MacGyver, or using this in the context of high school outreach programs), you're dealing with a less than ideal enzyme (error rate is terrible). Not really worth taking that chance in the research arena.
As far as MacGyvering other things, something like a plasmid prep would be easy, RNA work would be impossible, and microassays - well the first were actually prepared using ink jet printers, and before that it was simply a nylon membrane dotted by hand with probes.
Anyway, thanks for the plug for the Science Creative Quarterly. Glad it's reaching out.
If I were you Dr. Orac.
ReplyDeleteI would find out if the Russians (or Poles or Czech's maybe) are doing the very thing you are doing.
Odds are that if they are using Taq (?) that they have figured out a McGuyveresque solution for how to get it.
Necessity is mother of invention. Da.
Well, all of the molecular work I did, I did in another person's lab. His was a big lab (22 people at the time) with tons of money and letting me run a dozen gels or so was drop in his bucket. If it had worked, he would have had another easy paper to put his name on.
ReplyDeleteBack home, because I use a non-traditional animal model, I have had to, over the years, build almost all of my equipment myself. That's why I laughed at the Dremel joke the most (from the list of jokes I posted last night) - that's my favourite tool.
While becoming a biologist, I also became a carpenter, an electrician, and would have become a computer programmer if I was not fortunate to have a super-computer-geek for a technician for a while. All that trains one to be creative and not dependent on what suppliers offer - a good trait to have if one is a scientist.
Another thing you can save on is gel extraction kits.
ReplyDeleteYou can make "extraction columns" very cheaply and easily. You need a sponge, p1000 tips and microfuge tubes.
Take the sponge, cut into small pieces that can be stuffed into a p1000 tip, cut the tip to size so it fits in a microfuge tube. Close the cap, make some holes with a pin and autoclave. When you need to extract a band from an agarose gel, cut it out, place it on the sponge in the tip and centrifuge a few minutes at low rcf. The resulting liquid will have the DNA in the band.
The efficiency and reproducibility aren't very good, but it's worked for me in the past. I purified a PCR amplified band and managed to clone it ok.
RNA degradation is feared much too much. Exongenous RNase- clean the equipment. MiliiQ water-soap-mq-etoh-mq-meoh. Done.
ReplyDeleteDon't underestimate the power of decoy RNA or RNasin to reduce endogenous RNase activity. tRNA from Sigma will work well but do an organic extraction to get rid of any RNases that came along for the ride (diethyl ether is also necessary to get all the phenol out or else forget about spec'ing the sample). Reducing temperature will also slow down RNase activity.
Regarding the taq/coli protocol - you'll end up with RNases too after boiling.
There are legit strains of RNAP and taq that are his-tagged.
The final McGyver method would be to get a grad student or postdoc to write a grant using a commodore64. Mission accomplished.
A different POV from a more experienced PI.
ReplyDeleteI see a trackback (where excellent points are made)...but don't see how you get the ping.
We use a similar gel extraction technique in our laboratory:
ReplyDeleteTake 500uL tube, punch hole in the bottom using syringe. Stuff 1mm-2mm of glass wool in the bottom of the tube, insert 500uL tube into 1.5mL eppendorf. Put gel slice into 500uL tube, spin 10min, 6K in microfuge. Repeat if gel slice is large.
Do phenol-chloroform / precipitation afterwards.
I've had some success with *ligating* using the gel homogenate directly.